Ambient Proteins: Training Diffusion Models on Low Quality Structures
Published as an Spotlight in NeurIPS 2025 [Paper] [Code]
Authors: Giannis Daras*, Jeffrey Ouyang-Zhang*, Krithika Ravishankar, William Daspit, Costis Daskalakis, Qiang Liu, Adam Klivans, Daniel J. Diaz
Announcing Ambient Protein Diffusion, a state-of-the-art 17M-params generative model for protein structures.

Diversity improves by 91% and designability by 26% over previous 200M SOTA model for long proteins. The trick? We treat low pLDDT AlphaFold predictions as low-quality data.
Main Idea
Obtaining large structure datasets experimentally is impossible.
SOTA protein structure models are trained on AFDB (214M AlphaFold predicted structures) subsets.
AF accuracy drops with increasing protein length and complexity, making it hard to generate such proteins.
Ambient Protein Diffusion treats low pLDDT AF structures as low-quality data.
Instead of filtering them out (as done in prior work), we use them for a subset of the diffusion times.
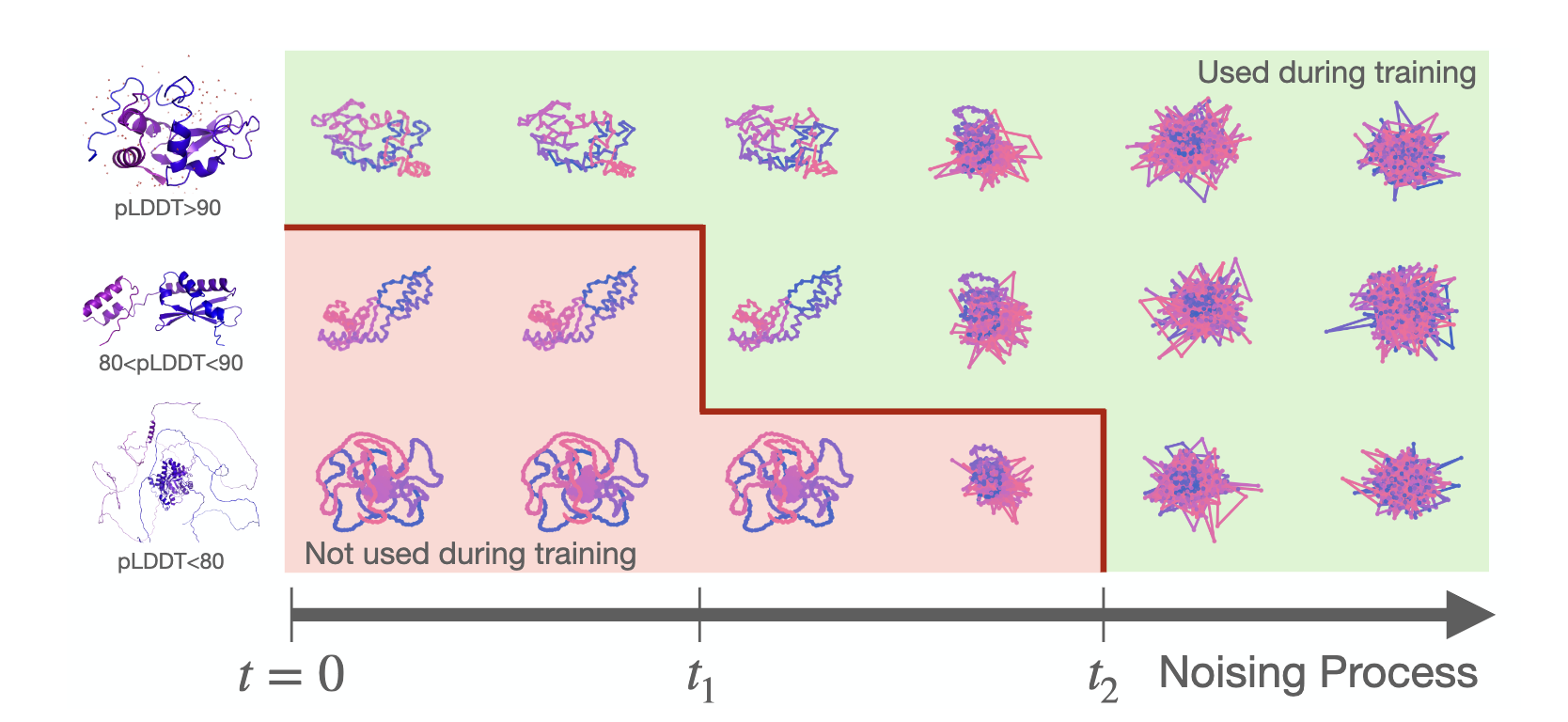
Enough noise “erases” the AF mistakes, and we can still learn from those structures.
Results
The results are quite strong.
Ambient Protein Diffusion substantially outperforms previous baselines in short and long protein generation.
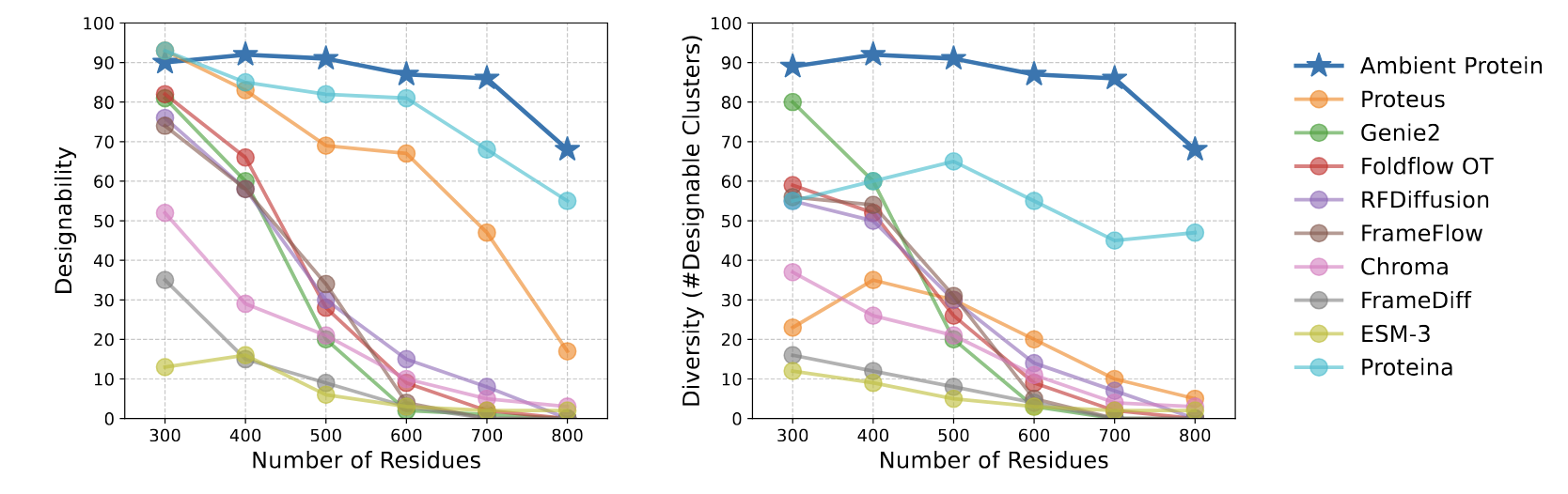
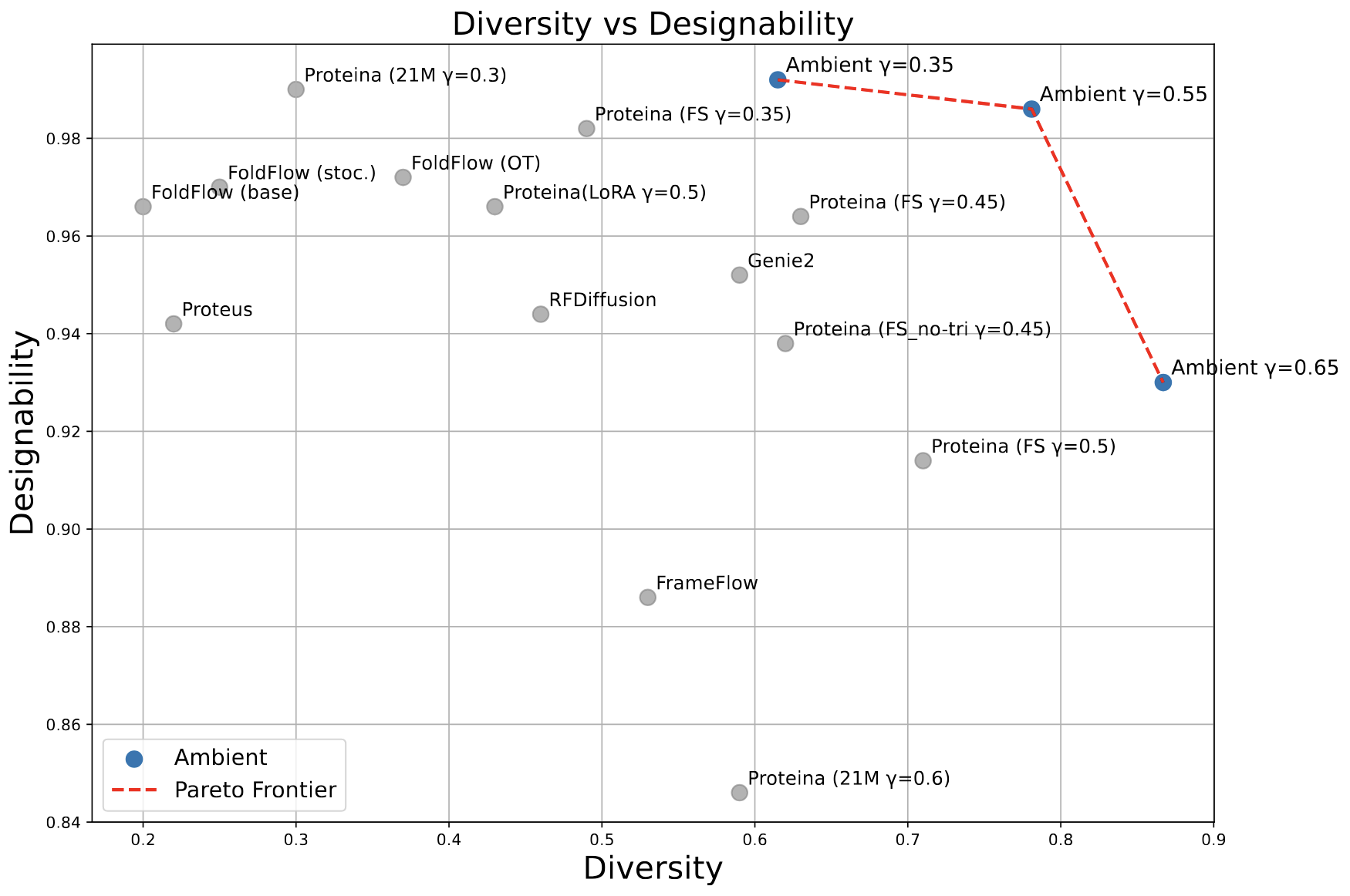
For short proteins, we dominate the Pareto frontier between designability and diversity, using a ~13x smaller model than previous SOTA.
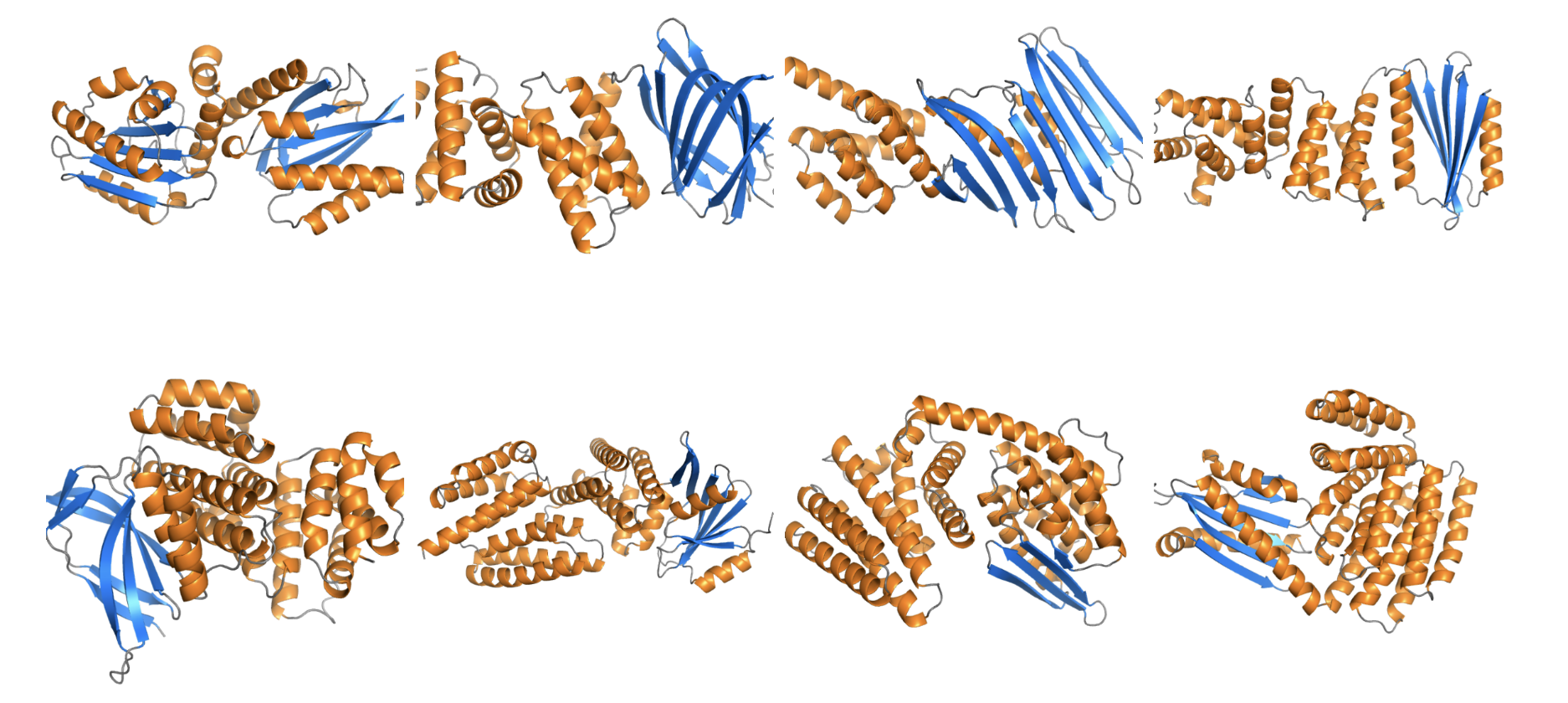
Here are some qualitative samples from our model:
Models
The following pre-trained Ambient Protein Diffusion models are available for download.
huggingface-cli download jozhang97/ambient-long --local-dir runs/ambient_long
huggingface-cli download jozhang97/ambient-short --local-dir runs/ambient_short
Detailed instructions on how to use the models can be found in the official code repository of the project.
Citation
If you use Ambient Protein Diffusion in your research, please cite our paper:
@article{daras2025ambient,
title={Ambient Proteins: Training Diffusion Models on Low Quality Structures},
author={Daras, Giannis and Ouyang-Zhang, Jeffrey and Ravishankar, Krithika and Daspit, William and Daskalakis, Costis and Liu, Qiang and Klivans, Adam and Diaz, Daniel J},
doi={10.1101/2025.07.03.663105},
journal={bioRxiv},
publisher={Cold Spring Harbor Laboratory},
year={2025}
}